Researcher Profiles

Adam Yala Ph.D.
University of California Berkeley
2023 Funding recipient
Machine learning to extract hidden insights from MDS bone marrow histology
Artificial Intelligence and Machine Learning in MDS 2023
PROJECT SUMMARY
CoPI with Alexander G. Bick, M.D., Ph.D., Vanderbilt University Medical Center
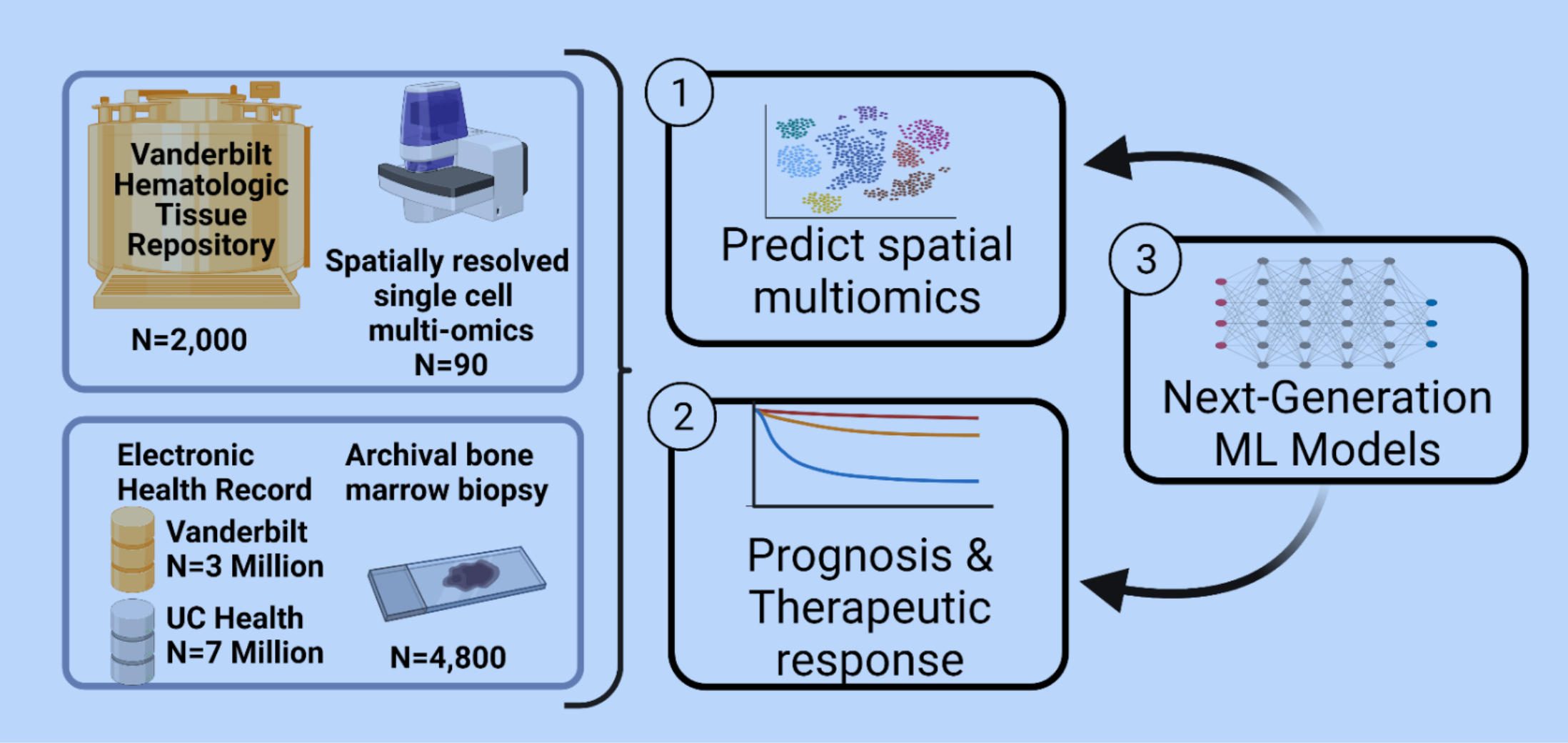
We seek to improve our understanding of myelodysplastic syndrome (MDS), a group of bone marrow disorders that affect blood cell production by using machine learning (ML) to reveal important patterns that are invisible to the human eye. Currently, doctors use bone marrow morphology (structure) to diagnose MDS by looking at bone marrow under a microscope. However,
We hypothesize that advanced ML models can extract important information from bone marrow microscope slides that is invisible to the human eye. We will use ML to enhance our understanding of MDS biology and improve patient prognosis
and treatment response. By combining these ML models with imaging data and large-scale measurements of single cells from MDS patients, we can better understand patient outcomes and identify new therapeutic targets.
Our project has three objectives. First, we will use ML techniques to predict single cell gene and multi-omic data from a simple histology slide. Second, we will use state-of-the-art ML models to classify patient disease prognosis and therapeutic response. Third, we will develop a new class of ML models that can analyze over 800x larger images than current methods, increasing the scale of data we can analyze with ML from micrometers to millimeters. These innovations will enable more accurate predictions for our first two aims.
To support our project, we will generate an open digital histology dataset of around 4,800 bone marrow biopsy samples and share this data with the community. We will also form a collaborative network with other funded teams to validate our results in other datasets. Our team is well prepared for this ambitious project and includes experts in MDS biology, AI/ML, and digital pathology.
Our research aims to benefit patients with MDS and the wider MDS research community by unlocking important insights from simple microscope slides. If successful, our algorithms will be able to recapitulate insights that can presently be identified only using very expensive spatial single cell experiments just for the ~$10 it costs to digitally scan a standard microscope slide. In short, our work would make personalized prognostication and treatment decisions widely accessible while answering important scientific questions about MDS biology.