Researcher Profiles

Vijay G. Sankaran, M.D., Ph.D.
2020 Funding recipient
Dissecting Pathogenic Mechanisms of ETV6 Deregulation in Myelodysplastic Syndromes
Discovery Research Grant 2020
PROJECT SUMMARY
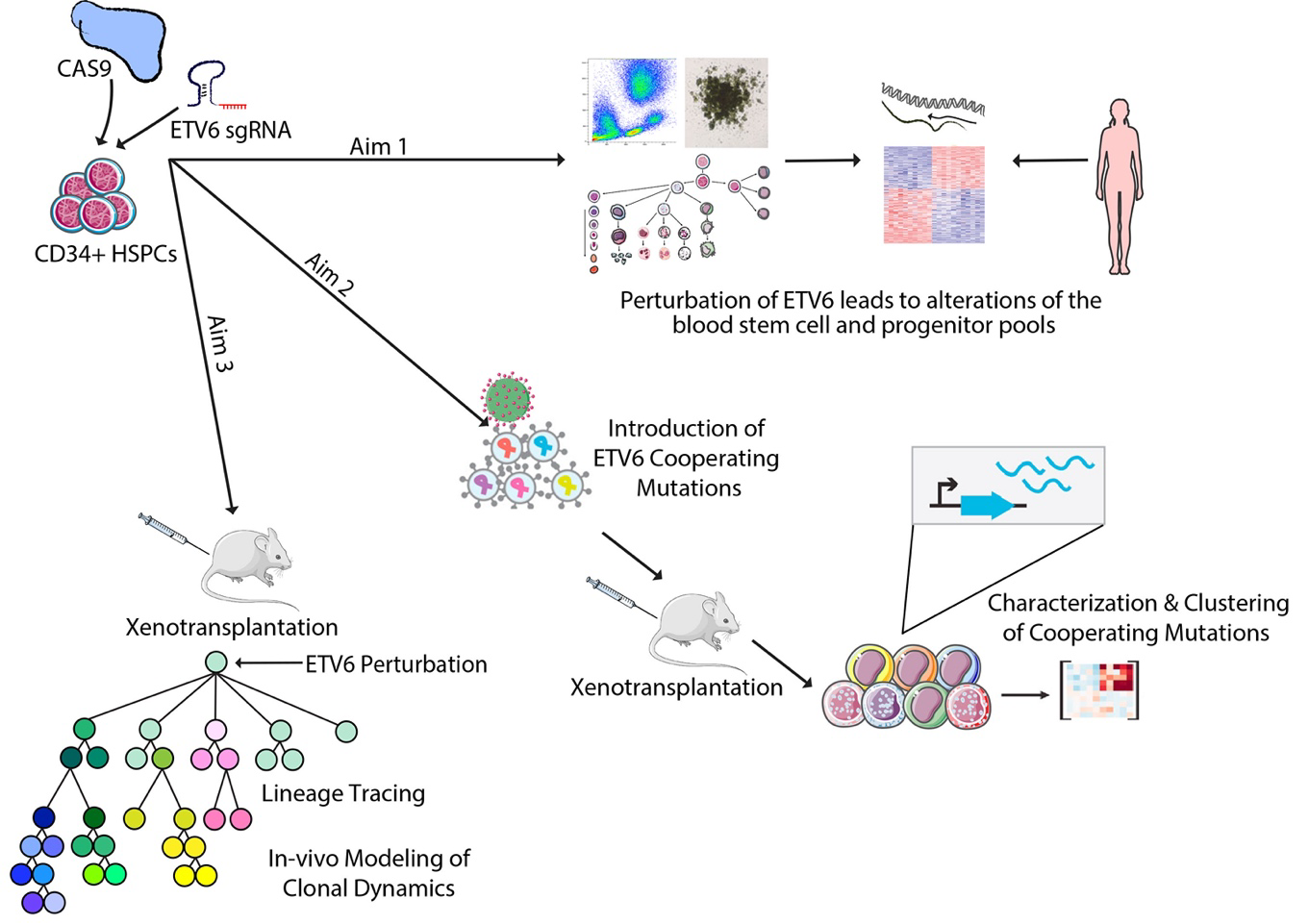
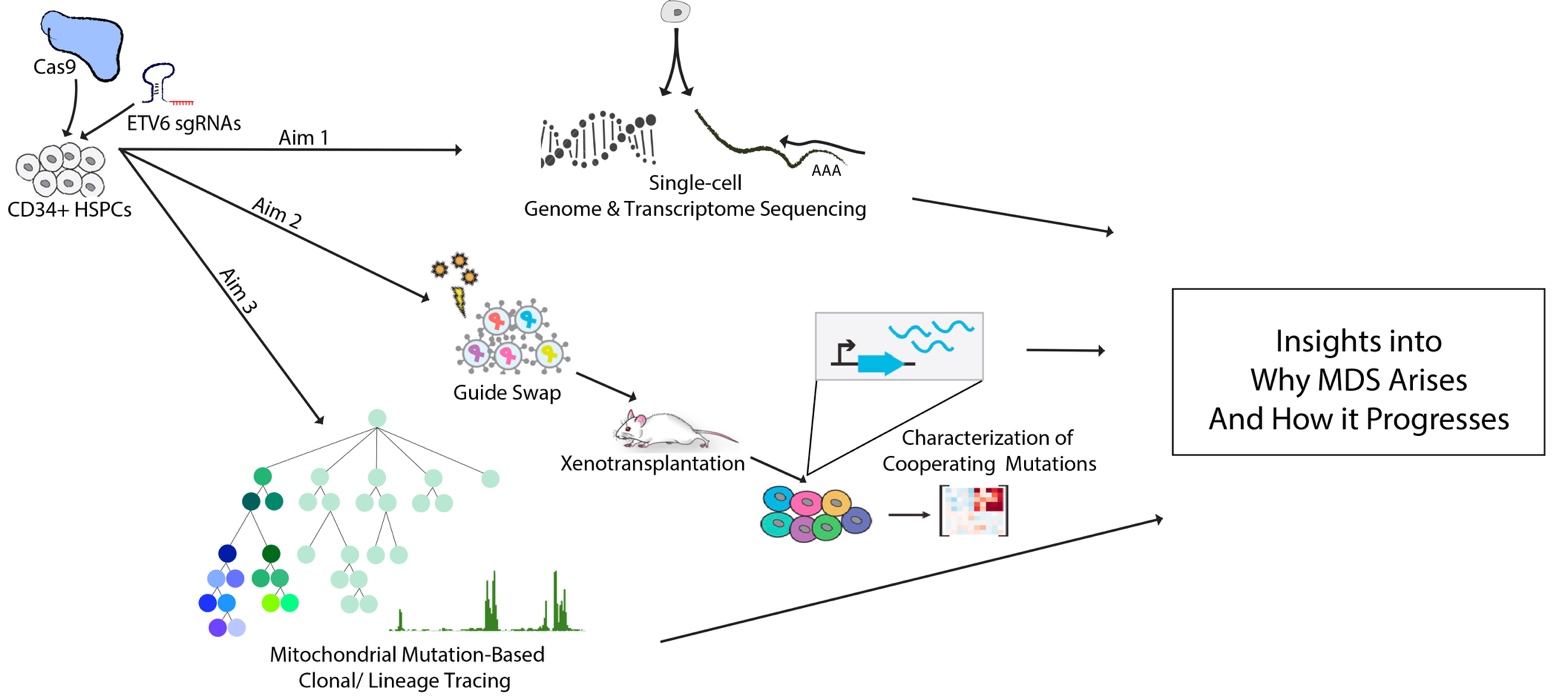
The objective of this ongoing study is to improve our knowledge of how inherited gene mutations play a role in myelodysplastic syndromes (MDS), both in predisposing to the disease and in progression of these disorders. MDS arises due to mutations in blood-forming stem cells that cause abnormal blood cell maturation. Recent studies have uncovered several recurrently mutated genes directly involved the development of MDS. A particularly intriguing gene in this group is ETV6, which encodes a transcription factor that plays a critical role in blood cell development and blood stem cell maintenance. Studying ETV6 holds promise, as it can drive MDS progression both in the form of inherited and acquired mutations, suggesting a central role for perturbation of this gene in the pathogenesis of MDS. Despite this, little is known about the mechanisms through which ETV6 impacts normal human blood cell maturation and how mutations in this gene can predispose to or drive the onset of MDS. This ongoing project includes a comprehensive set of studies to enable an improved understanding of the function of ETV6 in MDS. First, to gain insight into the function of ETV6 in human blood stem cells, we established a gene editing strategy to selectively recapitulate genetic mutations in this gene in stem cells derived from healthy donors. Employing such gene editing tools enabled us to examine the effects of mutations of ETV6 perturbation in an isolated and genetically tractable setting most relevant to what is observed in patients. Here, a number of intriguing observations were made. While a partial knock-down of ETV6 is overall well tolerated in human blood stem cells undergoing short term cell culture, highly effective knock-down of the gene could not be achieved in these cells. Although our genome editing strategy generally results in a highly effective knock-down of a given target gene, when targeting the ETV6 gene, mutations were introduced at only ~ 40-60% efficiency, across a number of different sites. Next, to determine the functional consequence of perturbations in this gene in human blood stem cells, we generated gene expression profiles in single cells using cutting-edge approaches. Intriguingly, we detected that perturbations of ETV6 in blood stem and progenitor cells alter gene expression profiles that lead to an enrichment of a specific subset of blood stem cells, a finding of interest in MDS pathogenesis, where mutated blood stem cells get altered to ultimately gain a competitive growth advantage over healthy cells. In addition, gene editing of the ETV6 gene leads to upregulation of pro-inflammatory pathways. This finding is of particular interest, as dysregulation of immune and inflammatory signaling is a well-known hallmark of MDS. To understand how blood cell formation is altered in this context, we next went on to characterize how mutations in ETV6 impact blood formation. For this, we subjected blood forming stem cells which underwent gene editing to multiple blood differentiation assays. Here, as a consequence of ETV6 perturbation we observed significant alterations of the composition of the pool of maturing blood cells, suggesting that mutations in ETV6 drives abnormal blood cell maturation. Intriguingly, upon serial sampling of differentiation cultures, the proportions of ETV6-edited cells were noted to significantly increase over time, from 40-60% at the beginning of culture, to 80-95% towards later time points. This may reflect a growth advantage of edited blood forming cells over unedited cells. Experiments to further examine the precise dynamics of how disruption of ETV6 affects maturing blood cells are ongoing. In future studies, we will subject bone marrow-derived blood cells from patients harboring ETV6 mutations to similar single-cell gene expression assays and compare the results of these two analyses. This will help delineate at which distinct stages of blood cell maturation ETV6 mutations act, and further define downstream pathways through which ETV6 impacts blood formation.
Patients with MDS often present with multiple disease-associated mutations affecting their blood stem cells. Furthermore, acquired ETV6 mutations have been established as a prognostic indicator of poor overall survival in patients with MDS. An ongoing aim of this project is to examine the clonal complexity of MDS. To further understand this, we will model how a variety of other mutations can act with ETV6 to cause MDS. For this, we have queried a local cohort of AML/MDS patients at our institution and obtained a list of gene mutations that commonly co-occur in the with ETV6 mutations. Using novel genome editing and screening strategies, we plan to leverage selective pressures from a known predisposing lesion in ETV6 on human blood forming stem cells to model clustering of such cooperating mutations. This will enable a more detailed understanding of whether and how certain mutations may cooperate, or interact directly, resulting in the heterogenous range of phenotypes along the MDS spectrum. We have recently significantly advanced our ability to carry out these screens by developing improved expression vectors that can also be detected with single-cell gene expression profiling. Finally, using an innovative lineage tracing approach that we have recently significantly advanced over prior approaches, we will dissect how disruption of ETV6 can alter the number of clones that compose the blood cell compartment. These experiments will provide additional insights into how ETV6 and other similar kinds of mutations can result in MDS progression. Continued funding from the Evans Foundation would enable us to continue these critical studies that should help improve our understanding of MDS predisposition and progression.