Researcher Profiles

Timothy M. Chlon, Ph.D.
Cincinnati Children's Hospital Medical Center
2023 Funding recipient
The Role of Missense DDX41 Mutations in MDS pathogenesis
Discovery Research Grant 2023
PROJECT SUMMARY
It is estimated that inherited predisposition accounts for up to 10% of cases of Myelodysplastic Syndrome (MDS) (10). The most common cause of predisposition to MDS are mutations in the gene DDX41. Unlike other mutations that predispose to MDS, DDX41 mutations do not cause a developmental disease, and they do not cause MDS in children or young adults. On the contrary, patients with these mutations typically live healthy lives until they are diagnosed with MDS or related diseases in the 5th decade of life or later. The DDX41 gene codes for a protein that has functions in essential cellular pathways including the splicing of RNA molecules and the synthesis of protein-producing complexes called ribosomes. Due to the importance of these processes for subsistence of cells, one functional copy of DDX41 is necessary to maintain normal cellular proliferation rate and survival (2). The most common inherited DDX41 mutations in MDS patients cause loss of the DDX41 protein. They do this by changing the number of nucleotides in the genetic code (insertion or deletion) or by changing the identify of one of the first three nucleotides, which are required for the start of protein production. Since the inherited mutations only affect one of the two copies of DDX41 the genome, it is thought that loss of one copy of DDX41 causes predisposition to MDS. However, up to 35% of the inherited DDX41 mutations observed in MDS patients do not cause loss of DDX41 protein. Rather, they change the identity of one of the 622 amino acids that make up the length of human DDX41 protein. In genetics, this type of mutation is called a missense mutation. Over 30 different missense DDX41 mutations have been observed in the germline DNA of MDS patients. However, the effect of these mutations on DDX41 function is almost entirely unknown. In this project, we aim to determine the effect of 20 of the most common missense DDX41 mutations observed in MDS on the function(s) of the protein. Knowing how these mutations effect DDX41 function will allow us to determine if these mutations affect blood cells in the same way as mutations that cause loss of DDX41 protein.
To test the effect of the missense mutations on the function of the DDX41 protein, we will engineer cells and strains of mice to express the mutant forms of the DDX41 protein that are observed in patients. We will use cultured cells in the lab to screen 20 of the most common missense mutations for their ability to function like normal DDX41. To do this, we will test the ability of each mutant DDX41 protein to maintain the proliferation and survival of cultured blood cells in the absence of normal DDX41. Remarkably, we have completed preliminary analysis of five different missense DDX41 mutations and found that all five mutant proteins possessed the required functions of DDX41 to support the proliferation and survival of cells. This finding suggests that missense mutations do not cause complete loss of DDX41 function and may be contributing to MDS by another mechanism. Importantly, DDX41 has other functions that are not essential to the proliferation and survival of cells. The best-studied of these is that it activates the innate immune system by sensing DNA that comes from bacteria or viruses that have infected cells. It is possible this function of DDX41 is involved in MDS since other proteins involved in innate immunity have been implicated in MDS. Thus, we will use a cellular system to test the effect of the missense mutations in DDX41 on the ability of the protein to activate the innate immune system. To do this, we will modify the genome of cells to have the missense DDX41 mutations and then treat these cells with a chemical that will make them turn into an innate immune system cell type called macrophages. Then we will test the ability of these macrophages to respond to a stimulus that mimics a bacterial infection. If the cells with DDX41 mutations fail to activate the innate immune system in comparison to cells with normal DDX41, then we will know that these mutations affect the innate immune system function of DDX41 and that this may play a role in MDS development in patients with these mutations. Finally, for three of the missense mutations that are most prevalent in patients, we will generate mice that have these mutations in their DDX41 genes and study how this affects their blood system. These studies will tell us if the mutations affect the blood system on their own, or if additional mutations in DDX41 are necessary to cause blood system defects, including MDS. One interesting facet of DDX41-mutant MDS is that many of the patients acquire a second DDX41 mutation in their blood cells when they develop MDS. Thus. we will generate mice that have both the inherited missense mutations and the second DDX41 mutation that occurs later. If these mice have defects in their blood system, we will study what blood cell types are affected by the mutations and how.
Altogether, these experiments will tell us which functions of DDX41 are important for MDS and how different DDX41 mutations affect these functions. Determining how different mutations predispose to MDS development is critical to developing successful treatment and prevention strategies for patients. The findings of our study have the potential to establish new classifications of DDX41 mutations that will inform treatment strategies based on DDX41 mutation status. For example, it is possible that some DDX41 mutations do not affect protein function at all and these patients’ treatment should not be influenced by DDX41 status, or that some DDX41 mutations promote MDS by a different mechanism than complete loss of DDX41 and that it is better to group these patients separately for disease treatment and prevention decisions.
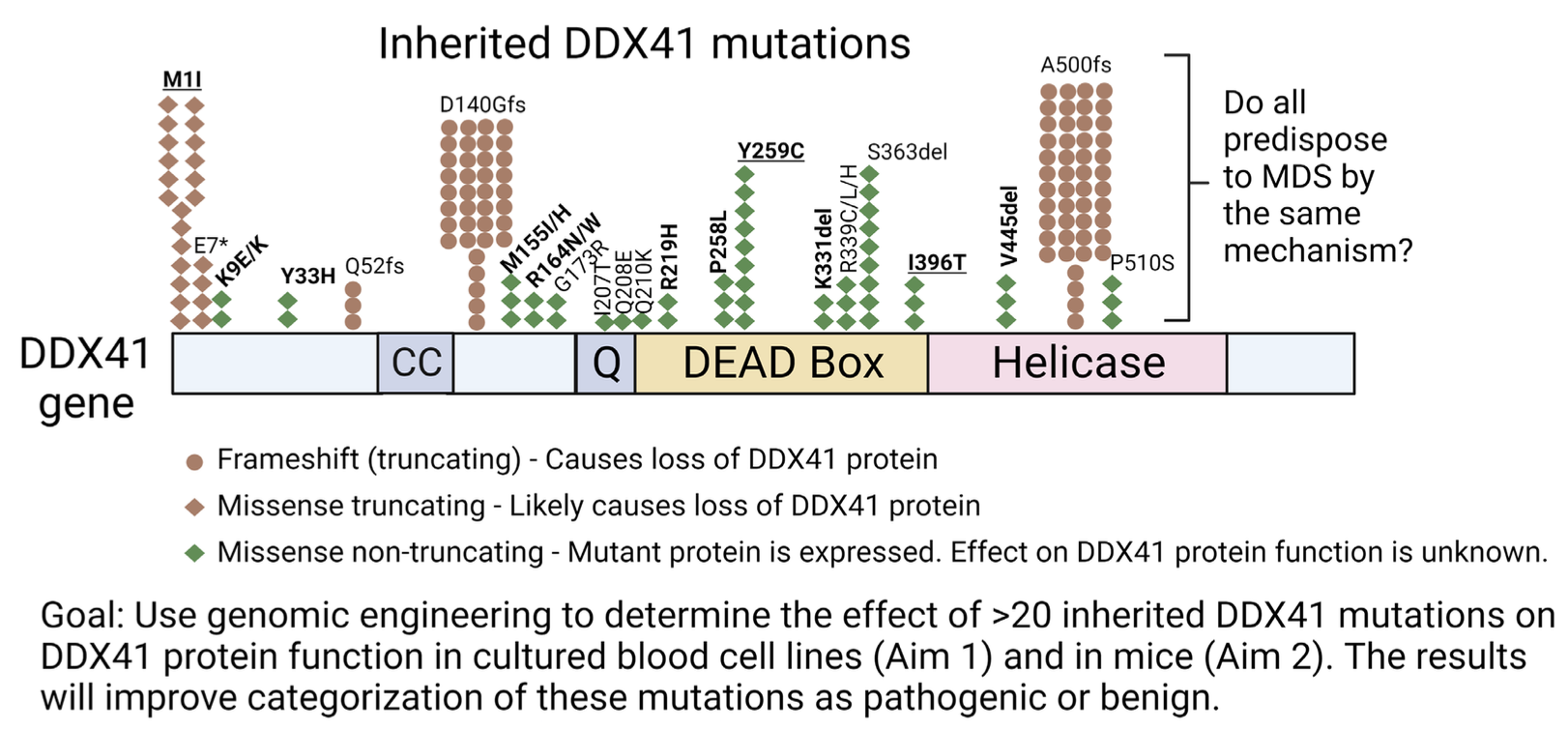 The figure above displays an approximation of relative frequency of mutations from Makishima et al Blood 2023 and Li et al Blood 2022. Many mutations that are rarely observed are not displayed for simplicity. The bolded and underlined missense mutations will be tested in mice and in two cell culture systems, the bolded will be tested in two cell culture systems, and the normal font will be tested in one culture system.
The figure above displays an approximation of relative frequency of mutations from Makishima et al Blood 2023 and Li et al Blood 2022. Many mutations that are rarely observed are not displayed for simplicity. The bolded and underlined missense mutations will be tested in mice and in two cell culture systems, the bolded will be tested in two cell culture systems, and the normal font will be tested in one culture system.
