Researcher Profiles

Prajwal C. Boddu, M.D.
2022 Funding recipient
Mechanisms of Altered RNA Fate and Processing in MDS-associated Mutations in Splicing Factors
EvansMDS Young Investigator Award
PROJECT SUMMARY
Mature blood cells arise from coordinated division and maturation of a few thousand blood stem cells. Diseases that affect this process lead to a failure to produce adequate number of functional blood cells, manifesting as anemia, infections, and bleeding. Myelodysplastic syndromes (MDS) are the most common cause of bone marrow failure in adults. Recent research has found that MDS is largely caused by acquired changes to the DNA (mutations) of blood stem cells affecting select groups of genes.
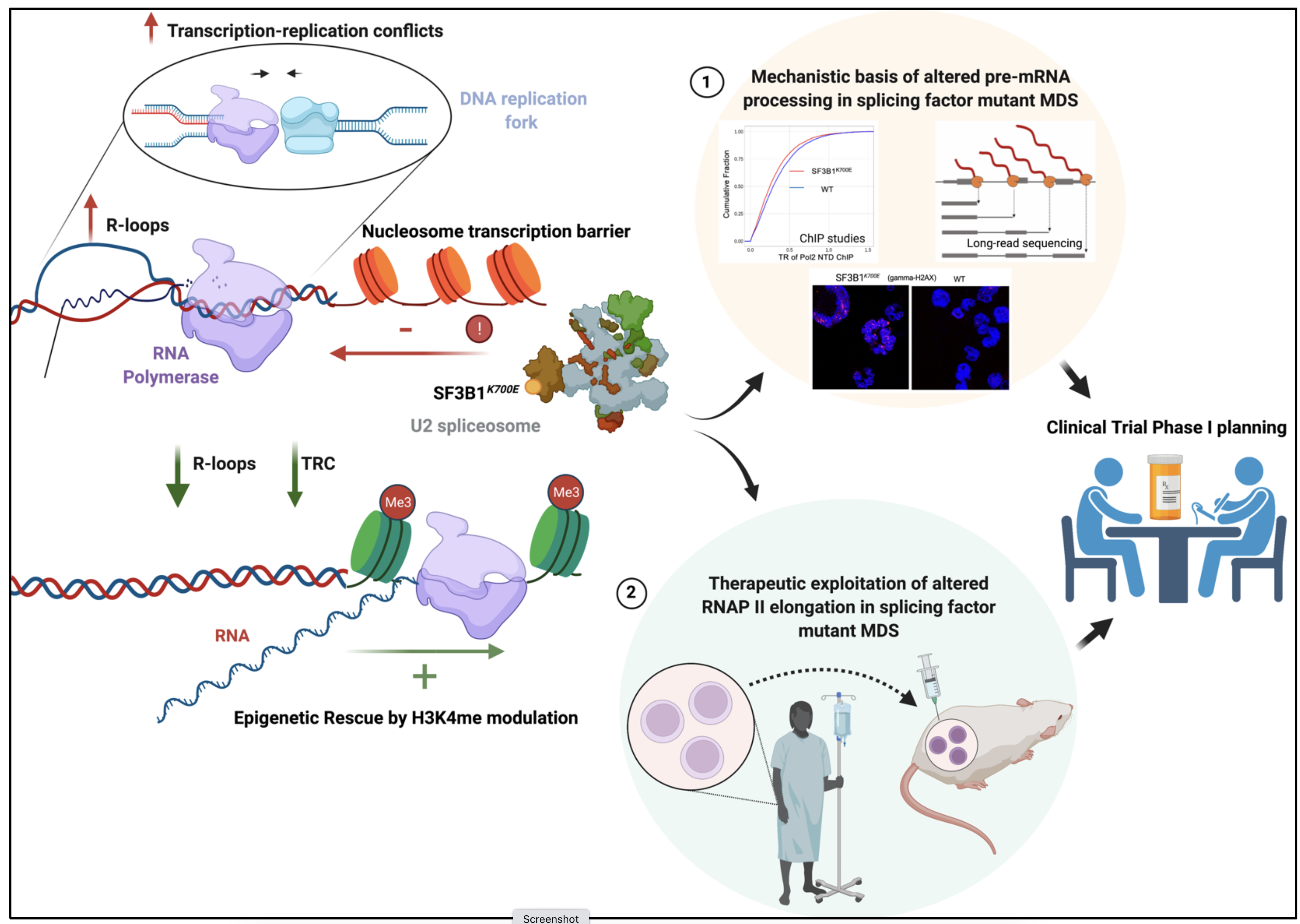
The most common group of genes, mutated in MDS, process ribonucleic acid (RNA) strands by cutting and rejoining them so they are suitable templates to make proteins. Called splicing factors (SFs), they are mutated in over half of all MDS. How such mutations lead to MDS is still an area of intensive investigation. Prior research has largely focused on changes to individual genes that are aberrantly processed by mutated splicing factors. Our preliminary results show that mutations in SF3B1 (the most frequently mutated SF) slow down the molecular complex that drives RNA transcription (called RNA Polymerase 2 or Pol2) from the DNA template. This leads to changes in the chromatin making the genome unstable and liable to acquire more mutations.
We propose that such a change to the speed of Pol2 transcription is the fundamental molecular defect that underlies MDS. Interestingly, our results also show that modulating expression of a group of genes called epigenetic regulators can correct Pol2 kinetics. This gives us an opportunity to therapeutically target SF mutations in MDS by modulating epigenetic genes. In this application, we seek to answer some fundamental questions regarding MDS: (1) How do mutations in SFs change Pol2 speed? (2) How does modulating certain epigenetic proteins help cells overcome the Pol2 speed defect? (3) Can we exploit these insights for therapeutic benefit? For these studies, we have generated novel genome-edited models suitable for large-scale biochemical investigations. We will also utilize blood stem cells isolated from MDS patients and mouse models of MDS wherein patient samples are engrafted into mice with compromised immune systems (patient derived xenografts).
Our studies will provide fundamental insights into the mechanisms by which mutations in SF lead to MDS. It will also inform novel approaches to therapeutically target the mutations by modulating epigenetic factors.